The goal of this project is to develop heterogenous parallel program to accelerate molecular docking simulation. Some features include;
- AutoDock 4 implementation for semiempirical energy function.
- Historical genetic algorithm for conformational search.
- Python implementation using OpenCL as the accelerator.
Documents:
- NTU Dissertation
- NTU Dissertation Slides
- Paper for IEEE Life Sciences Grand Challenges Conference 2013 in Singapore
- Poster for IEEE Life Sciences Grand Challenges Conference 2013 in Singapore
Tested on:
- Python 2.7.3
- NumPy 1.7.1
- PyOpenCL 2013.1
To run:
- Go to PyNeuroDock directory
- Execute python NeuroDock.py
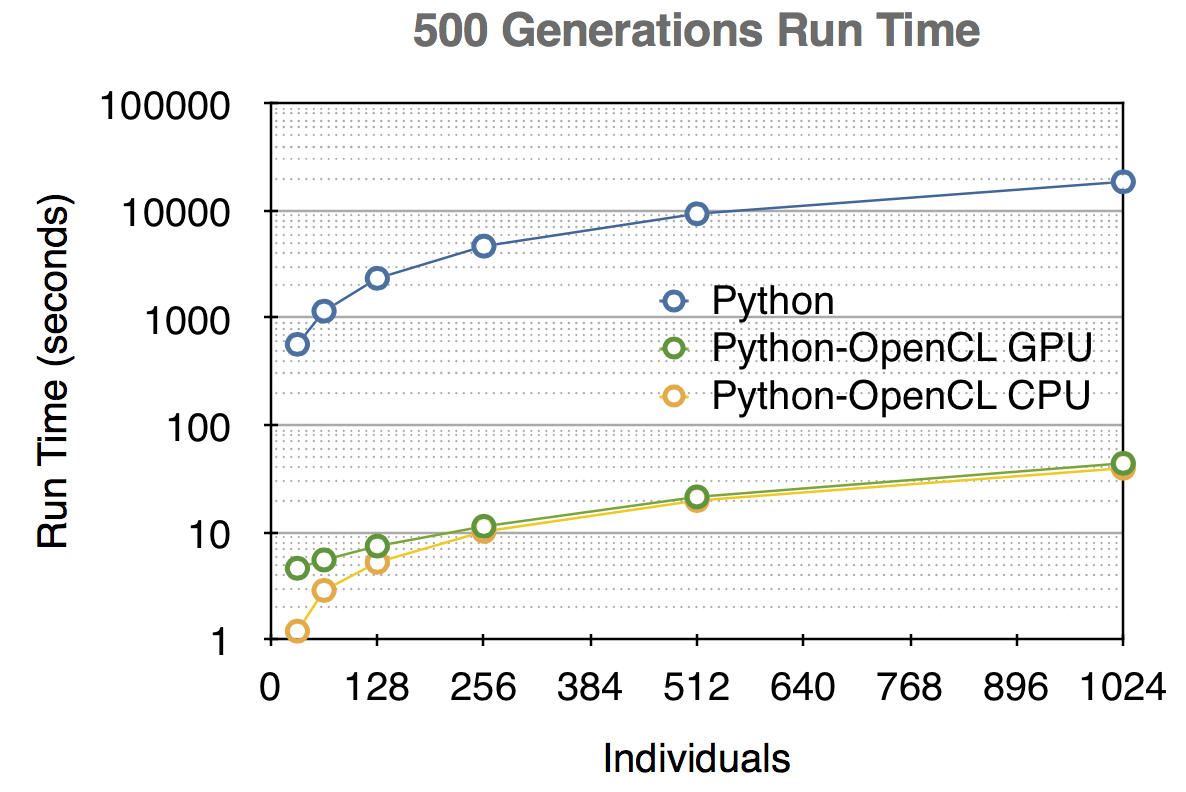
Benchmark:
- On Unix shell (Bash), PyNeuroDock directory, execute: ./Benchmark.sh
- Python: Sequential processing run on 2.3GHz Intel Core i7 (1 thread)
- Python-OpenCL GPU: Parallel processing run on NVIDIA GeForce GT 650M 1GB (384 CUDA cores)
- Python-OpenCL CPU: Parallel processing run on 2.3GHz Intel Core i7 (4 cores, 8 threads)